- Quantification of the strength of noncovalent bonds
- Binding strength and selectivity of chemotherapeutical drugs Description: Drugs used for chemotherapy are usually considered to have low specificity, which means any given drug may bind with multiple nucleic acid sequences without much difference. Using FIRMS and SURFS techniques, we are able to precisely determine the binding strength of drugs to nucleic acids. By defining the differential binding force, which equals the binding force of drug-DNA complex subtracting that of DNA alone, we are able to map the binding specificity of drugs. Interestingly, we revealed a mutual selectivity between DNA sequences and drug chirality. We are currently attempting to correlate force measurements with thermodynamic parameters to facilitate drug screening. In addition, we are expanding the method for drugs targeting protein-protein and protein-nucleic acid interactions.
- Frameshifting during ribosomal translocation (in collaboration with Prof. Wang,
- The role of mechanical force in biology (in collaboration with Prof. Wang,
- Low-field magnetic resonance imaging
Description: Noncovalent bonding is the primary type of chemical interactions in biology. Compared to covalent bonds, noncovalent bonds are usually weak and reversible. However, quantification of the bonding strength remains challenging. We have invented a unique force spectroscopy that combines mechanical force and magnetic detection. The first generation of this technique is force-induced remnant magnetization spectroscopy (FIRMS), which offers 1.8 pN force resolution. Recently, we developed the second generation of this technique by using acoustic radiation force and automated atomic magnetometer. The technique is termed as super-resolution force spectroscopy (SURFS) that reaches 0.4 pN force resolution. Noncovalent bonds can be distinguished down to a single hydrogen bond difference.
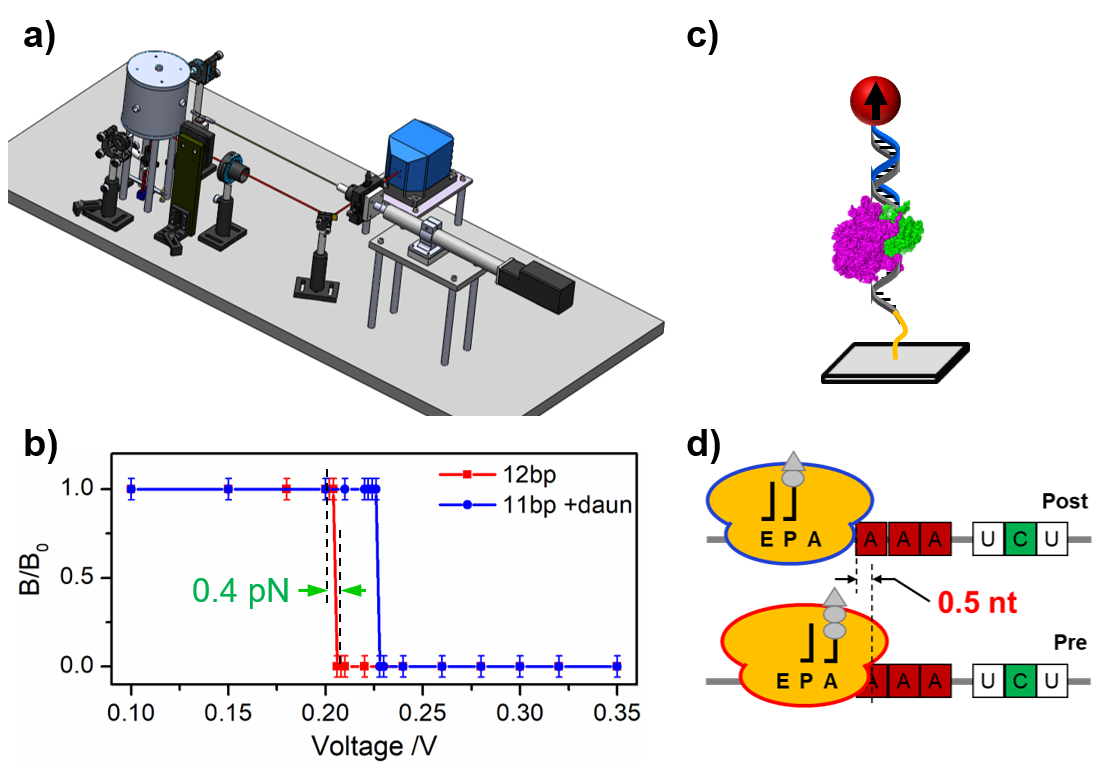
Representative publications:
Yao, L. & Xu, S.-J. Force-induced remnant magnetization spectroscopy for specific magnetic imaging of molecules. Angew. Chem. Int. Ed. 50, 4407-4409 (2011).
Chen, Y.-T., Jamison, A. C., Lee, T. R. & Xu, S.-J. Quantitatively resolving ligand-receptor bonds on cell surface using force-induced remnant magnetization spectroscopy. ACS Cent. Sci. 2, 75-79 (2016).
Jia, H., Wang, Y. & Xu, S.-J. Super-resolution force spectroscopy reveals ribosomal motion at sub-nucleotide steps. Chem. Commun. 54, 5883-5886 (2018).
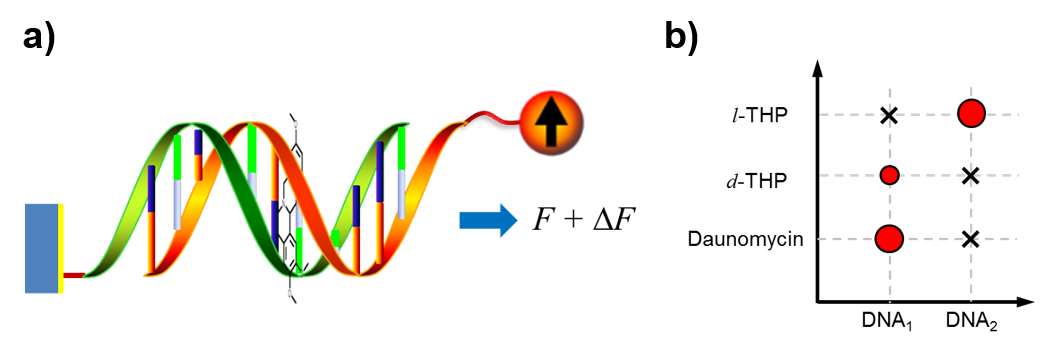
Representative publications:
De Silva, L., Yao, L., Wang, Y. & Xu, S.-J. Well-defined and sequence-specific noncovalent binding forces of DNA. J. Phys. Chem. B 117, 7554-7558 (2013).
Hu, Q. & Xu, S.-J. Sequence and chiral selectivity of DNA-drug interactions revealed by force spectroscopy. Angew. Chem. Int. Ed. 53, 14135-14138 (2014).
https://www.uh.edu/nsm/biology-biochemistry/people/profiles/yuhong-wang/)
Description: Frameshifting refers to the ribosome translocates other than the normal 3-nucleotide step along the mRNA. It is commonly encountered in disease-related slippery sequences. Depending on the relative steps compared to normal translocation, frameshifting can be categorized as -2, -1, +1, +2 frameshifting, in which the ribosome moves 1, 2, 4, and 5 nucleotides, respectively. Because the adjacent reading frames differ by only one nucleotide, our force spectroscopy techniques are uniquely suited to distinguish different frames. Using FIRMS, we have identified a novel -2 frameshifting on the GA7G motif. Recently using SURFS, we revealed that the ribosome moved by half a nucleotide from the post-translocation state to the pre-translocation state of the next cycle. More detailed work and the effect of antibiotics are under investigation.
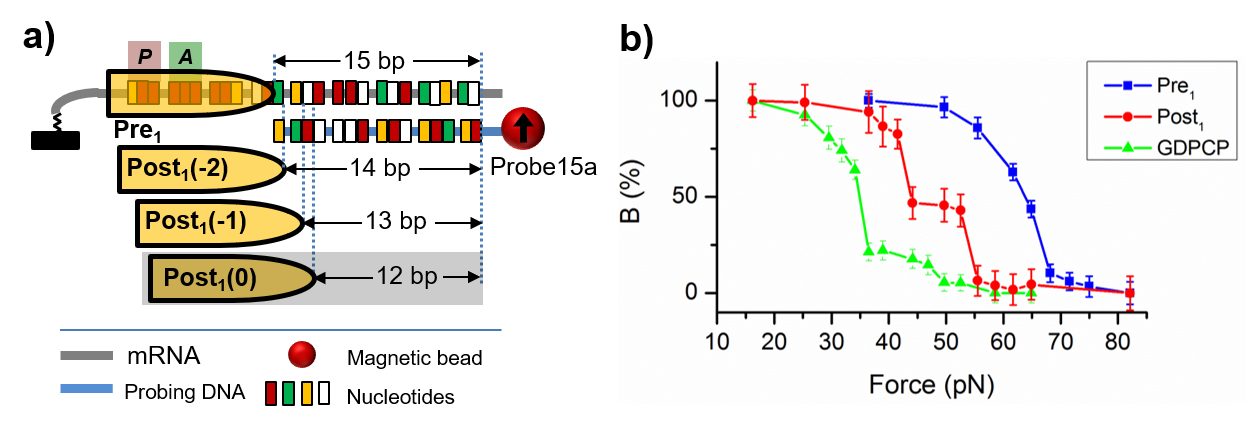
Representative publications:
Tsai, T.-W., Yang, H., Yin, H., Xu, S.-J. & Wang, Y. High-efficiency “-1” and “-2” ribosomal frameshiftings revealed by force spectroscopy. ACS Chem. Biol. 12, 1629-1635 (2017).
Jia, H., Wang, Y. & Xu, S.-J. Super-resolution force spectroscopy reveals ribosomal motion at sub-nucleotide steps. Chem. Commun. 54, 5883-5886 (2018).
https://www.uh.edu/nsm/biology-biochemistry/people/profiles/yuhong-wang/)
Description: Mechanical forces of substantial amplitudes may be the driving factor for biological processes involving significant molecular motions. Due to the lack of suitable techniques, there are arguments on their roles and amplitudes. One particularly important example is the ribosomal translocation—is it force directed or Brownian motion governed or both? We used a series of DNA duplexes formed between the probing DNAs and the mRNA to serve as internal force calibrator. The dissociation percentages of the duplexes indicated that there was an 89 pN mechanical force generated by the elongation factor G (EF-G). This is the most direct approach to determine the EF-G power stroke. Such force can only be generated by the GTP hydrolysis. More systematic work is currently ongoing.
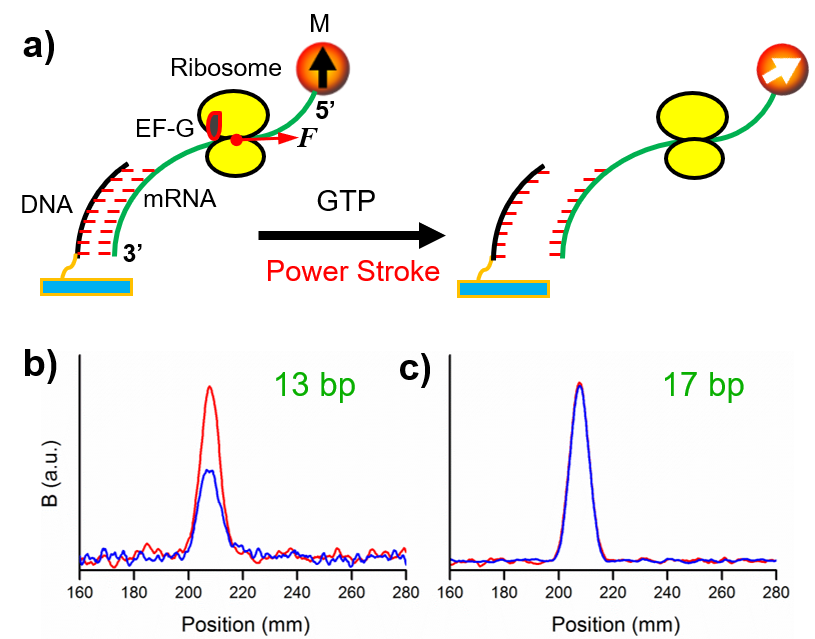
Representative publications:
Yao, L., Li, Y., Tsai, T.-W., Xu, S.-J. & Wang, Y. Noninvasive measurement of the mechanical force generated by motor protein EF-G during ribosome translocation. Angew. Chem. Int. Ed. 52, 14041-14044 (2013).
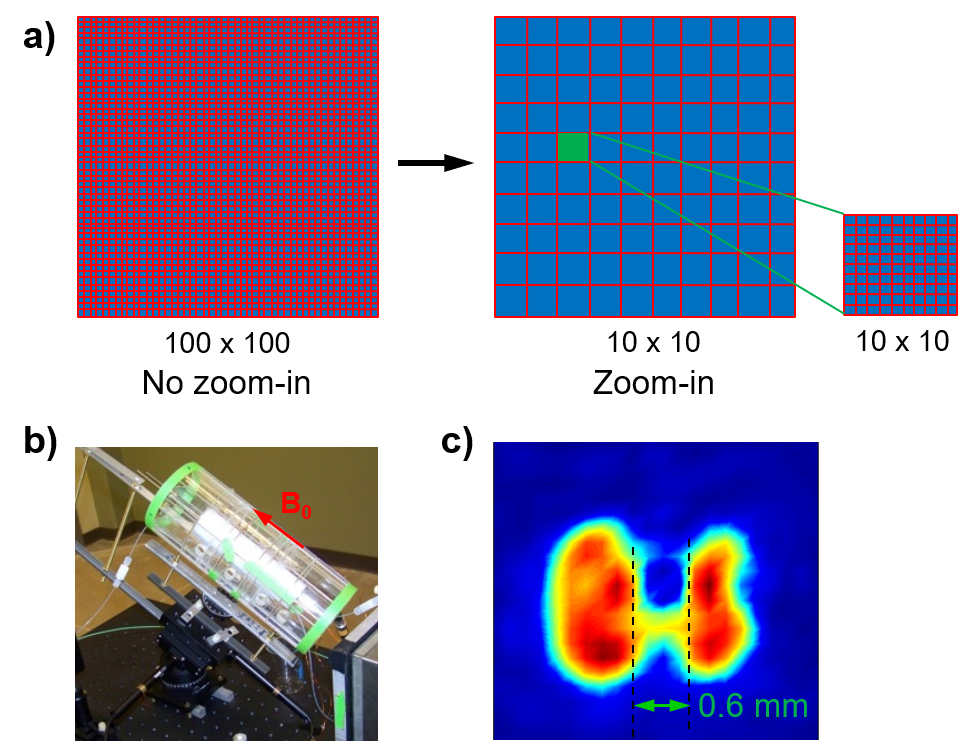
Description: Magnetic resonance imaging (MRI) is one of the most powerful medical imaging modalities. There are, however, certain scenarios that conventional MRI is not applicable, for example, for patients with metallic implants or with claustrophobia. In addition, the high cost of MRI scanners sometimes are note available, for example, in remote area or space traveling in the future. These inapplicable conditions are the motivations for developing low-field MRI. By using an atomic magnetometer to directly detect the nuclear magnetization, it is possible to obtain MR images in a low magnetic field such as the Earth’s magnetic field. We used an innovative zoom-in scheme that achieved sub-millimeter resolution. Further development is needed in order to improve both the feasibility and spatial resolution.
Representative publications:
Ruangchaithaweesuk, S., Yu, D. S., Garcia, N. C., Yao, L. & Xu, S.-J. Applications of optically detected MRI for enhanced contrast and penetration in metal. J. Magn. Reson. 223, 20-24 (2012).
Ruangchaithaweesuk, S., Chintamsetti, V., Yao, L., Tsai, T.-W. & Xu, S.-J. High-resolution optically-detected magnetic resonance imaging in an ambient magnetic field. J. Magn. Reson. 233, 1-6 (2013).

